Transform hours of tedious manual data analysis into minutes with connected scientific data and AI
Like many modern biotech companies, Sail Biomedicines performs multiple types of experiments. Their platform, which enables AI-based programming of endless RNA™ (eRNA™), leverages multiple workflows to deeply characterize RNA. But to do so, the team faces the challenge of combining and normalizing results and metadata across multiple data systems. For Sail, two of these systems include Benchling and AWS:
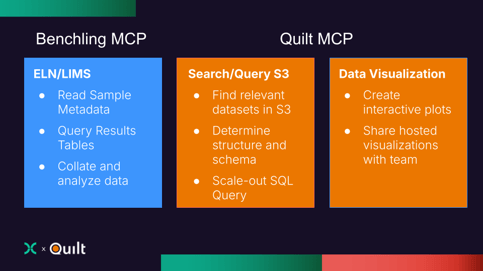
- Benchling: The system of record, which tracks sample registration, assays, and experimental context, and is home to low-throughput experimental results (ELISAs, qPCR, Flow Cytometry, etc.)
- AWS: The home for high-throughput data sets, including large next-generation DNA sequencing-based results.
Even answering simple questions requires data from both systems. For example, if a scientist wants to know, “Can I use RNA expression levels of eRNA™ constructs from a specific program as a proxy for ranking protein expression?”, they would have to:
- Find the relevant datasets in Benchling and AWS
- Pull expression data from Nextflow results in Amazon S3
- Match the metadata with results in the Benchling warehouse
- Collate the data
- Perform the analysis and generate a report
This process can take a significant amount of time, adding friction to the design, build, and test cycle of improving eRNA™ performance. When scientists are stuck waiting to answer “Did my experiment work and what should I do next?” innovation slows.
How AI and Model Context Protocol (MCP) Enable Faster Scientific Data Analysis
At the 2025 Nextflow Summit, Vasisht Tadigotla from Sail Biomedicines presented a new, AI-first approach that replaces these time-consuming, manual steps with a single AI prompt. Connecting Anthropic's Claude Sonnet 4.5 to MCP servers from Benchling and Quilt.bio (the scientific data management system for AWS), bench scientists can get answers to their cross-assay questions in minutes instead of hours.
Giving LLMs the Tools They Need for Scientific Data Analysis

Today's LLMs can reason about scientific questions, but their context windows are much smaller than biotech datasets. For an LLM to help scientists analyze data across systems, a single AI session must search and query data from multiple sources, and therefore relies on numerous tools:
- Searching and filtering massive datasets.
- Querying structured tables (SQL over columnar data).
- Joining data across disparate systems (e.g., Bench metadata ↔ Pipeline outputs).
- Executing complex computations.
- Producing reproducible artifacts (versioned datasets, plots).
That’s where MCP comes in. MCP is a standard that allows Large Language Models (LLMs) to interact with external tools in a structured, auditable way. Instead of each system having its own isolated AI, MCP allows one AI session to securely access multiple systems through standardized tool interfaces. Crucially, this is achieved without the model ingesting all the data. Data stays in its source system, and access control is enforced by that system, which makes MCP servers well-suited to highly regulated, large-scale scientific environments.
For companies – like Sail – that manage large-scale scientific data in AWS, AI requires an extra layer to transform raw S3 (records) into analysis-ready data. Quilt provides this layer. With Quilt, users can store, organize, and retrieve terabyte-scale data effortlessly within Amazon S3 and attach it to Benchling Notebooks via seamless integration. This integration ensures that all data is securely managed, easily accessible, and fully labeled for streamlined workflows, enhancing productivity in the lab.
MCP Tools Needed to Bridge Benchling and AWS
A Concrete Example: RNA–Protein Correlation at Sail Biomedicines
Sail scientists wanted to understand how RNA expression (on S3) for eRNA™ constructs in a specific program correlated with protein abundance in Benchling.
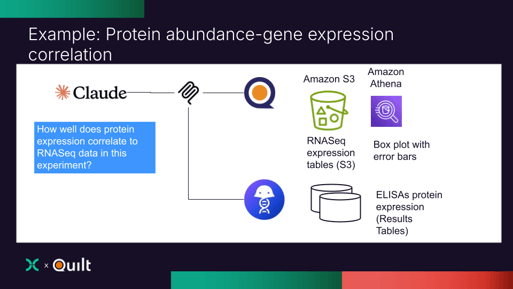
Using Anthropic's Claude Sonnet 4.5 connected to MCP Servers from Quilt and Benchling, Sail scientists ran a single prompt that:
- Located relevant Quilt packages in S3
- Queried RNA-seq expression tables at scale
- Pulled matching protein assay results from Benchling
- Aligned samples and gene identifiers
- Generated comparative visualizations
- Packaged the analysis as a new Quilt package
Total time to analysis: ~10 minutes
No file wrangling.
No hand-written SQL.
No hand-off to a data science team.
With GenAI and MCP bridging their Benchling and AWS systems, Sail’s Bench scientists can now:
- Perform meaningful analyses themselves
- Quickly see computational results in their experimental context
- Iterate faster on hypotheses, accelerating experimentation
Data and platform teams can:
- Avoid bespoke integrations
- Preserve AWS-native governance
- Focus on advanced analysis rather than routine requests
MCP enables AI systems to accelerate scientific workflows that cut across system boundaries. Data analysis that normally requires hours to days of tedious, manual work across bench scientists and computational teams takes minutes with this breakthrough approach. To learn more, watch the video:
Comments